Overview
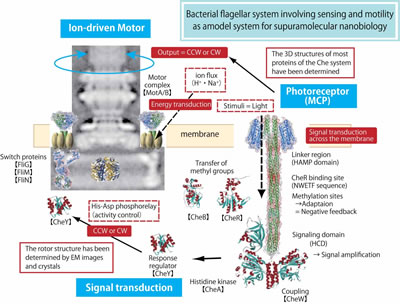
If you think about bacteria, what comes to mind? Some might say that bacteria are bad organisms that cause infectious diseases, others may say that fermented soy beans (Natto) are made using bacteria. These are true statements, but bacteria are actually very useful in modern life science technology. Such a small organism can move by itself (motility), and can sense environmental information (recognition). Bacteria have the ability to move toward better environments (taxis). Our research interests focus on energy transduction and sensory transduction in bacteria. A special emphasis is put on the supramolecular complexes located in the cytoplasmic membrane, namely the flagellar motor and the photoreceptor-kinase complex. The bacterial flagellar motor is an elaborate molecular machine driven by ion (proton or sodium ion) motive force. We want to understand how electrochemical energy (i.e. ion flux) is converted to mechanical energy (i.e. torque) to drive this tiny rotary motor. The photoreceptor-kinase complex is a polymodal molecular sensor that regulates the flagellar motor to achieve taxis. We want to understand how an light signal is received and transmitted by a receptor to produce an intracellular signal and how it is canceled by a feedback regulatory system. Furthermore, we want to know how the shape and function of the biological membrane are regulated using giant liposomes. Our group focus on the membrane functions and studied them by the aspects of system biology by use of structural biology, nanotechnology, molecular biology, molecular genetics, biophysics, and so on. |
Mechanism of energy coupling in the flagellar motor
The bacterial flagellum is a helical filament driven by a reversible rotary motor at its base. The energy source for the motor is an electrochemical proton (or sodium ion) gradient across the cytoplasmic membrane. The flagellar filament is a huge organelle, whose length is several times larger than the cell body. This corresponds to the case that one who is 160 cm tall swings a string of 4 m long at 1,700 rpm, that speed is close equivalent to the F1 car engine! This flagellar system is the only rotary locomotive organelle to be described. However, the mechanism underlying flagellar motor rotation has yet to be fully elucidated. Therefore, we are trying to clarify the energy coupling using the following approaches: (i) Transform proton-driven motors into sodium-driven motors by genetic manipulation, (ii) Precisely measure flagellar rotation using a tiny bead attached to the rotating filament, (iii) Investigate behavior of the stator proteins on the membrane by using GFP-fusion constructs, (iv) Determine the protein structure of the motor, and (v) Reconstitute the entire motor in a proteoliposome using purified motor components. With these projects, we wish to elucidate the rotation mechanism of this magnificent molecular machine. |
Regulation of the number and placement of the polar flagellum in Vibrio
Precise regulation of the number and placement of flagella is critical for the mono-flagellated bacterium Vibrio alginolyticus to swim efficiently. We have previously proposed a model in which a putative GTPase of FlhF localization at the pole determines the polar location and generation of a flagellum, a putative ATPase FlhG interacts with FlhF to prevent FlhF from localizing at the pole, and thus FlhG negatively regulates the flagellar number in V. alginolyticus cells. By clarifing the function of FlhF and FlhG, we would like to know how to precisely regulate the number and placement of flagella. |
Molecular mechanisms of circadian rhythms
We aim to understand how organisms adapt to daily environmental alteration. Living organisms have evolved a cellular clock mechanism of approximately 24-hour period (the circadian clock). To understand the molecular mechanisms of the circadian clock and how it makes the lives of organisms efficient, we have been studying cyanobacteria, the simplest organism to have the circadian clock. Using forward molecular genetic approaches, three clock genes, kaiA, kaiB, and kaiC, that encode a central component of the cyanobacterial clock were identified in 1997. Surprisingly, circadian oscillation can be reconstituted in the test tube by mixing the three proteins and ATP. We would like to address the molecular mechanisms that generate the oscillation and how the phosphorylation cycle regulates the cellular metabolism. As cyanobacteria are simple prokayotes, studies of the circadian clock can be more easily advanced than in most of the higher organisms. |
References
|
1. Sowa, Y., et al. (2014). Proc. Natl. Acad. Sci. USA. 111(9):3436-4341. |